Chapter Leads: Harold Lehmann, Lisa Eskenazi
7.1 Introduction
The purpose of this chapter is to introduce the core building blocks for performing analyses in the N3C Data Enclave (see The N3C Data Enclave and Data Access in the Introduction).
We start with the Observational Medical Outcomes Partnership (OMOP) vocabulary, since all data are represented in the OMOP common data model (CDM) format. This chapter will describe the difference between the OMOP concept id unique identifier and the source value contributed by a site, the notion of hierarchy across concepts, and the OHDSI tools outside the Enclave that can help. Because we are working in a secure NCATS Enclave environment, we cannot rely on OHDSI tools outside the Enclave, so knowing the basic applications available within the Enclave and how these data are organized is crucial for the proper conduct of research.
We next present the notion and management of concept sets. These are concept ids that are taken as synonymous, for a given research purpose, and have their own set of tools.
We turn next to constructs – derived variables and facts – derived from concept sets and the raw data, including N3C OMOP additions that build on the OMOP data.
We round out this discussion with an introduction to external datasets that have been brought into the Enclave and the process to bring further datasets in.
Later chapters will give more details on how to use the resources and tools described here.
7.2 OHDSI Basics
This Observational Health Data Sciences and Informatics (OHDSI) basics section introduces the OHDSI community and provides an introduction to OMOP vocabularies, and vocabulary search tools such as ATHENA and ATLAS. For further OHDSI collaborative knowledge and exploration, refer to The Book of OHDSI which serves as a central knowledge repository (OHDSI, 2019). The Book of OHDSI is a living document, community-maintained through open-source development tools, and evolves continuously. The online version, available for free, always represents the latest version.
7.2.1 OHDSI Community
OHDSI is an international, interdisciplinary research collaborative that promotes open-source research. Its purpose is to create open-source solutions that bring out the value of observational health data through large-scale analytics. OHDSI has established an international network of researchers and observational health databases with a central coordinating center housed at Columbia University. More information about OHDSI can be found at https://www.OHDSI.org.
7.2.2 OMOP Vocabulary
The Book of OHDSI states:
The OMOP Standardized Vocabularies, often referred to simply as “the Vocabulary”, are a foundational part of the OHDSI research network, and an integral part of the CDM. They allow standardization of methods, definitions and results by defining the content of the data, paving the way for true remote (behind the firewall) network research and analytics. Usually, finding and interpreting the content of observational healthcare data, whether it is structured data using coding schemes or laid down in free text, is passed all the way through to the researcher, who is faced with a myriad of different ways to describe clinical events. OHDSI requires harmonization not only to a standardized format, but also to a rigorous standard content.
N3C relies heavily on the OMOP common data model which is part of the heart of the N3C harmonization process. OMOP, along with other data partners from ACT, TriNetX, and PCORI, ingest their data feeds into a central OMOP N3C repository that is then available within the N3C Data Enclave for researchers.
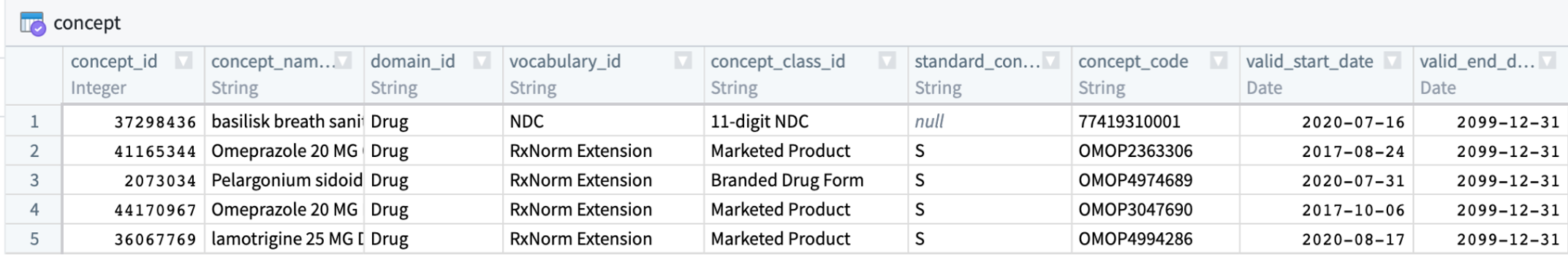
The OMOP CDM is an open-source, community standard for observational healthcare data and consists of clinical tables describing conditions and events from patient records and vocabulary tables describing clinical concepts. The central table in the OMOP vocabulary system is the table, concept.
7.2.2.1 Vocabulary Concepts
The meaning of every data element in the OMOP vocabulary, and many of the values of the data themselves, are represented by codes in the source data that get mapped to OMOP concept ids. Suppose a physician diagnoses a patient with diabetes and types “I10” into the EHR (which uses the ICD-10 vocabulary). The “I10” value is stored in OMOP as the source_value, and is then translated via the OMOP Vocabulary to the standard concept id, “320128”.1 N3C analyses use standard concept ids, as opposed to specific codes that aren’t necessarily consistent across hospitals.
The Book of OHDSI has a detailed chapter on Standardized Vocabularies. The OHDSI vocabularies are in a standardized CDM structure that houses existing vocabularies used in the public domain. This CDM compiles standards from disparate public and private sources as well as some OMOP-grown concepts.
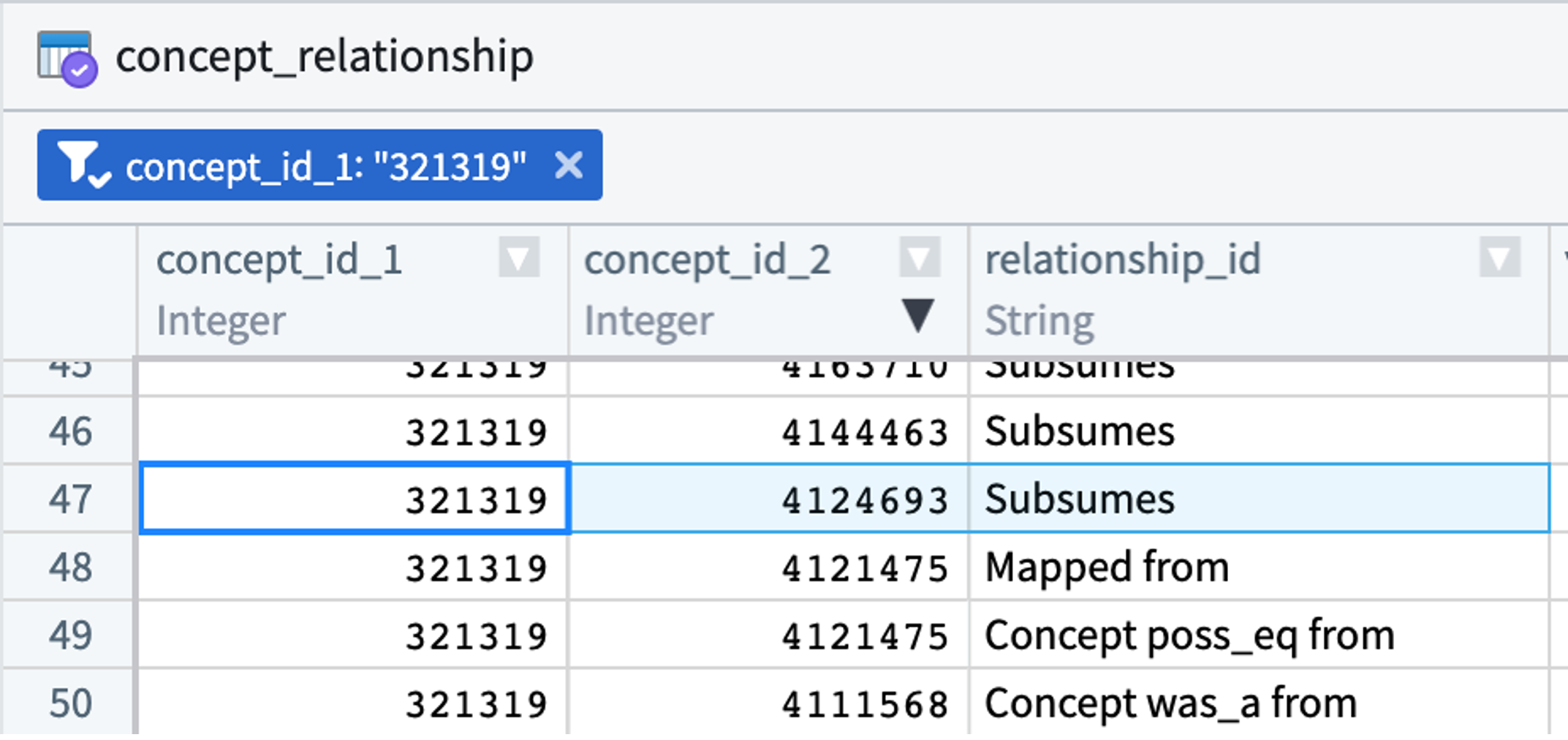
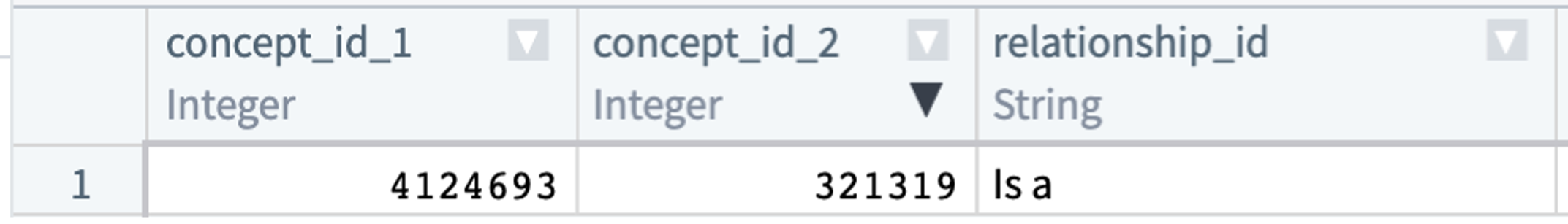
Concepts are related to each other. For instance, hypertrophic cardiomyopathy (concept id 4124693) is a type of cardiomyopathy (concept id 321319). The concept_relationship table stores these relationships. While analysts are welcome to use this table in their queries, there are other approaches to doing the same. (See concept sets.)
Every relationship type connecting two OMOP concepts has a converse. In the concept_relationship table, this means a parent-child relationship will be represented (redundantly) by two records. Hypertrophic cardiomyopathy (concept id 4124693) is a type of cardiomyopathy (concept id 321319):
The hierarchical relationship between the two concepts is represented once (above) with a “Subsumes” record and again (below) with an “Is a” record. concept_relationship contains only direct relationships between pairs of concepts. For hierarchical relationships, more distant relationships are found in the concept_ancestor table.
The concept_relationship table contains many other relationship types beyond hierarchical relationships: “Mapped from”, “Maps to”, “Has brand name of”, “Consists of”, etc.
ATHENA is an OHDSI tool for exploring OMOP vocabularies and even downloading them; its primary use in N3C is for looking up individual terms. More details on using ATHENA to search terms are documented on OHDSI’s publicly-viewable GitHub repository. ATLAS is an OHDSI tool to explore concepts and their relationships. See the section below on concept sets that describes further uses of the ATLAS tool.
The OMOP vocabulary is updated regularly to keep up with the continual evolution of sources. Its vocabulary maintenance and improvement is an ongoing activity that requires community participation and support. Releases and release notes are published via Git Releases on the OMOP Vocabulary GitHub repository. The most recent specification documentation can be found at OMOP Common Data Model. New versions of the OMOP vocabulary are downloaded monthly by the Enclave managers.
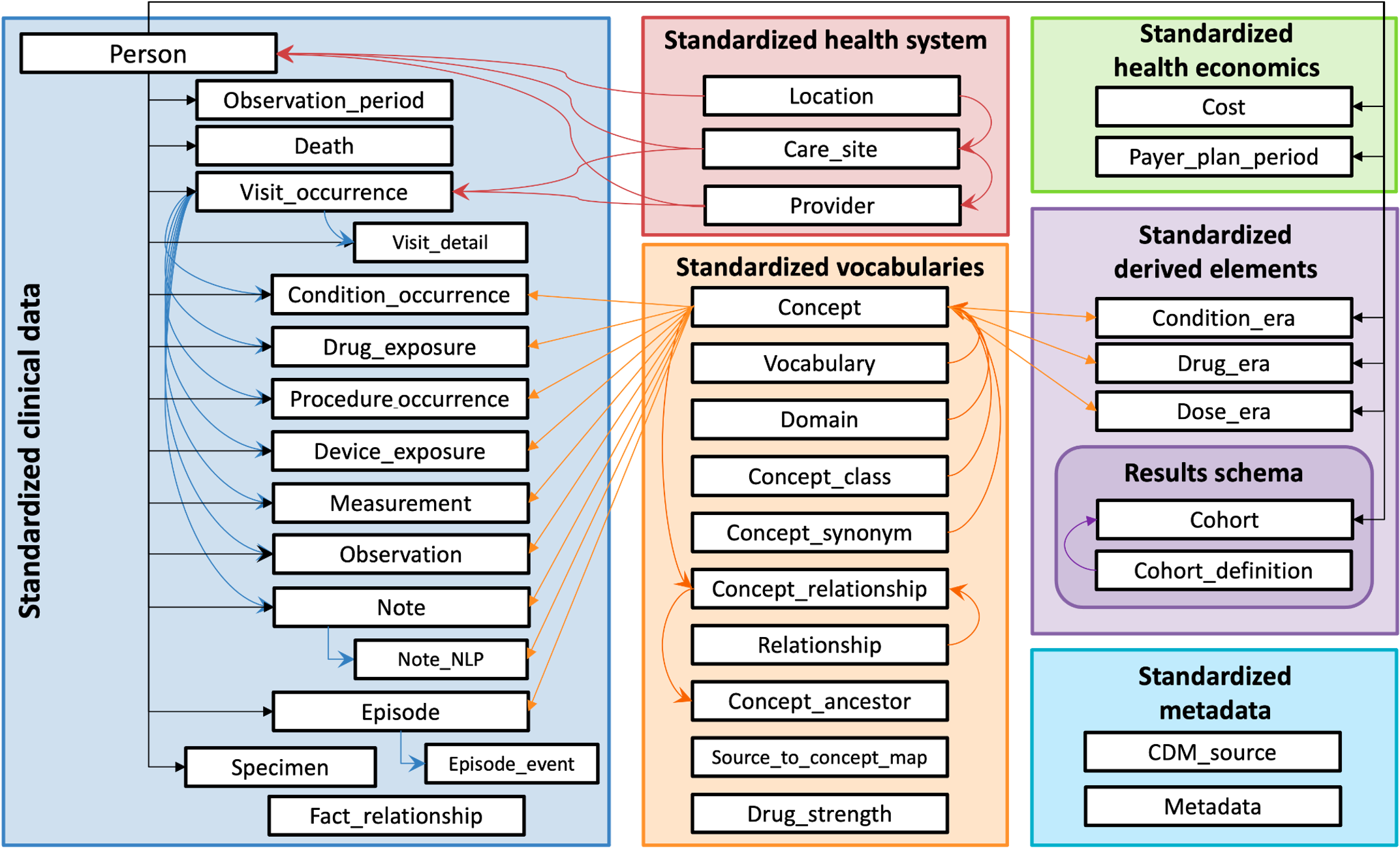
The OMOP CDM assembles these relationships in the Standardized Vocabulary section of the CDM. These tables are available to N3C researchers, along with the Standardized clinical data tables that store the patients’ data (expressed using the concept ids). Data are available as well in the Standardized health system, Payer_plan_period (not cost), and Standardized derived elements tables.
To learn more about OMOP and get acquainted with the CDM used in the Enclave and its functionality, see OMOP 101: A Crash Course in OMOP Standard Vocabulary and Chapter 11 in this guide.
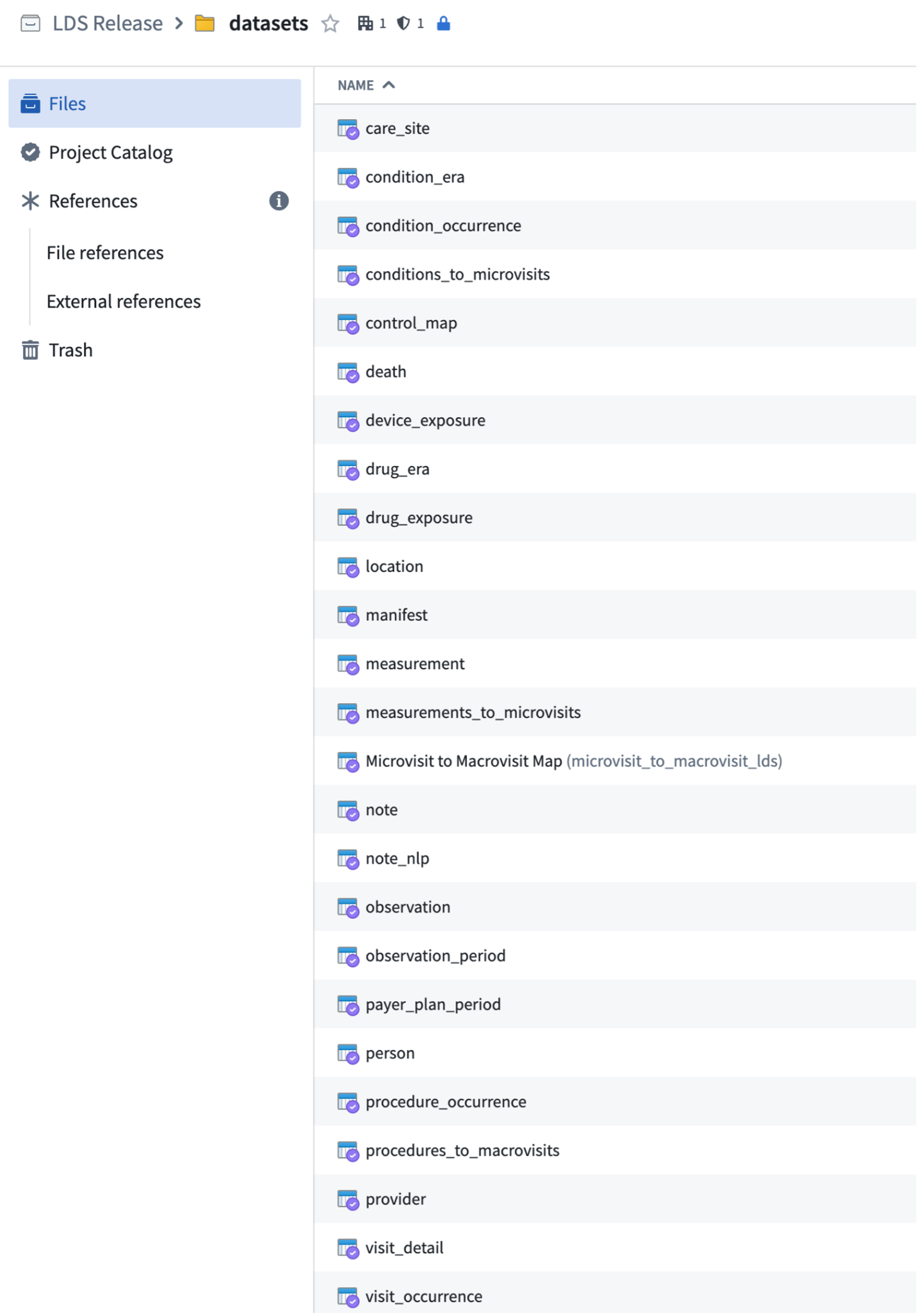
The figure below shows all OMOP tables within the Enclave.
7.2.2.2 Domains
Tables may contain data from more than one “domain”. There are currently 36 Domains in OMOP; the 5 primary clinical Domains are Drug, Condition, Observation, Procedure, and Measurement, plus others. They classify concepts in the concept table.
7.2.2.3 Vocabulary Updates
As stated above, source vocabularies change. For instance, at the start of the pandemic, there were no ICD-10 codes for “COVID-19 infection”, and therefore, there were no OMOP vocabulary codes for it and, therefore, no N3C codes. When we need a code, but there is none in the OMOP vocabulary, we create a local code. When the OMOP vocabulary team releases a newer, relevant code to replace the local code, the Enclave attempts to contact everyone who has employed the local code, notifying them of the change.
More subtle issues are when the source vocabulary managers change or decommission existing codes or add new codes (that do not have a local code equivalent), and the OMOP managers incorporate those changes in the vocabularies brought into the Enclave every month. Another subtlety is when the relationships between codes are changed. The Enclave attempts to contact those who have used codes related to the changed code. Generally, the analyst has used those codes in concept sets, to which topic we now turn.
7.3 Concept Sets
Any study or analysis performed in the N3C Data Enclave will start with identifying meaningful clinical conditions and events in patient data. We attempt to determine the presence or absence of clinical phenomena in patient history through the presence or absence of certain concept codes in patient records (Gold et al., 2018, 2021). For any given phenomenon used in an analysis, however, a single concept code will seldom be sufficient, and concept sets are used.
There are many reasons to specify a concept set:
- Variables in your study are implemented using concept sets.
- Having dozens or hundreds of concept ids in a single concept set makes analysis much easier.
- It makes the analysis easier to peer review.
- It makes your research coherent with the research of others who use the same concept set(s).
- Chunking concept-ids together into concept sets with different names and intentions makes your own (research) intentions clearer.
If you are publishing your results, concept sets are considered a publicly viewable part of your work. Such public viewing is important.
- It is required by the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for publishing observational research.
- Open Science expects reproducibility, and reproducibility in EHR-based research requires knowing how variables (concept sets) were defined.
- It enables other N3C researchers to use your concept sets, which is what makes research coherent across N3C projects.
Concept sets that are “finalized” within N3C will have information about them posted to our publicly-viewable GitHub repository. Such posting is part of the process known as making research FAIR (findable, accessible, interoperable, reusable). This repository is functionally our Shared Variable Library. Other variable definitions beyond concept sets are posted there (formulas, harmonized values, cohort definitions).
The following concept set content will be made public in Zenodo and/or GitHub:
- Concept set name, version number, date of finalization
- Intention
- Limitations (of the concept-ids within the concept set)
- Issues (with respect to data in the Enclave, so analysts are forewarned)
- Provenance (how the concept set was assembled, which communicates how trustworthy the concept set is)
- Attributions (who did what)
- The list of codes (in JavaScript Object Notation (JSON) format, which can be imported directly into Observational Health Data Sciences & Informatics (OHDSI)’s Atlas tool)
- Zenodo persistent document object identifier (DOI)
7.3.1 What is a concept set?
The Book of OHDSI explains concept sets as:
A concept set is an expression representing a list of concepts that can be used as a reusable component in various analyses. It can be thought of as a standardized, computer-executable equivalent of the code lists often used in observational studies. A concept set expression consists of a list of concepts with the following attributes:
- Exclude: Exclude this concept (and any of its descendants if selected) from the concept set.
- Descendants: Consider not only this concept, but also all of its descendants.
- Mapped: Allow to search for non-standard concepts.
While ATLAS concept sets may contain standard and non-standard concepts, Enclave concept sets may include “standard” (SNOMED-based) concepts only. Thus, the “Mapped” attribute is rarely helpful, as it applies only if using source data.
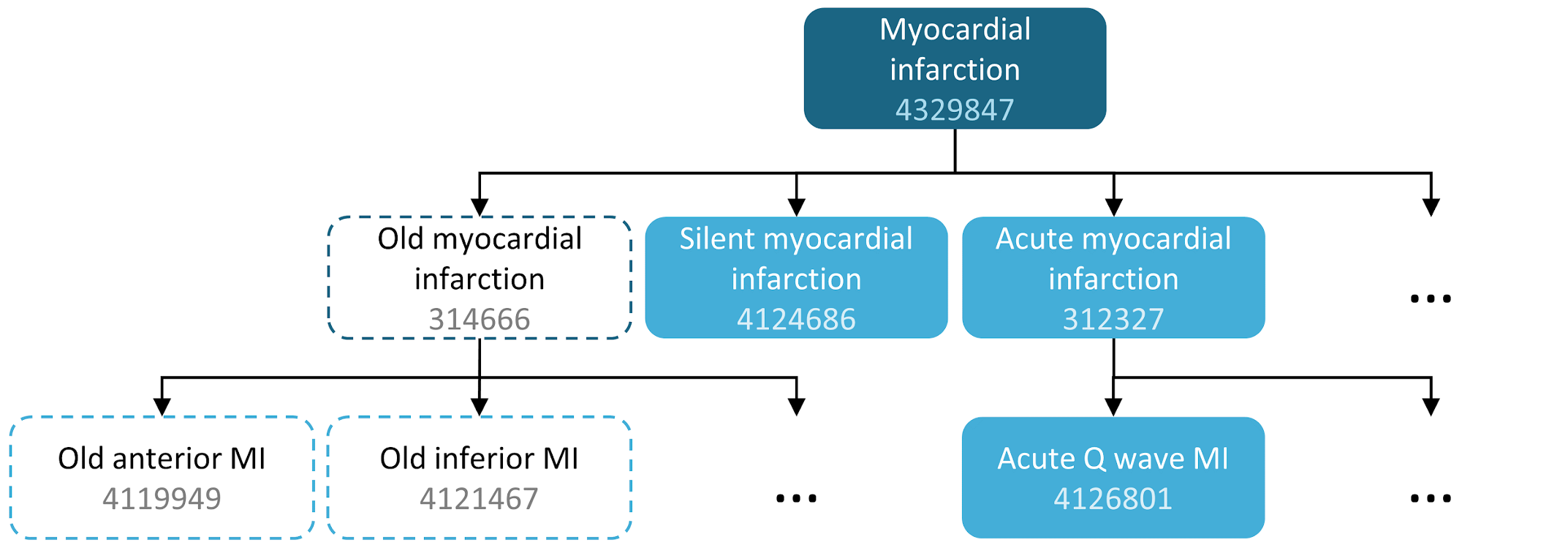
For example, a concept set expression could contain two concepts as depicted in The Book of OHDSI’s Cohort chapter. Here we include concept 4329847 (“Myocardial infarction”) and all of its descendants, but exclude concept 314666 (“Old myocardial infarction”) and all of its descendants.
| Concept Id | Concept Name | Excluded | Descendants | Mapped |
|---|---|---|---|---|
| 4329847 | Myocardial infarction | NO | YES | NO |
| 314666 | Old myocardial infarction | YES | YES | NO |
As shown in Table 7.1, this will include “Myocardial infarction” and all of its descendants except “Old myocardial infarction” and its descendants. In total, this concept set expression implies nearly a hundred Standard Concepts. These Standard Concepts in turn reflect hundreds of source codes (e.g., ICD-9 and ICD-10 codes) that may appear in the various databases.
ATLAS is used by the OHDSI community to construct concept sets. Best practice is for each research institution to have its own CDM instance. The Enclave, for technical reasons, cannot implement ATLAS. In the future, N3C hopes to provide a public tool but it is unclear when this release will occur. Users without their own local ATLAS may explore its utility using a publicly available one on OHDSI.org. Do beware all public tools are intended to be a demonstration site to showcase the power of the open-source tool stack. They are not intended for active research projects. These deployments are not running on a real OMOP CDM. They use synthetic data like the CMS Synthetic Public Use Files (SYNPUF) in the OMOP CDM format. The vocabulary schema will reflect whatever version of the vocabulary was in place when that OMOP CDM ETL was run. It will not reflect the most recent version of the OMOP vocabulary, which may be problematic for research projects that use newly approved vaccines or medicines. You should not make the habit of saving content within the public instance. It can be reset and wiped at any point in time. If you enjoy using ATLAS, consider spinning up your own instance via Broadsea, OHDSI in a box, or other OHDSI Community resources to jump-start users with their own ATLAS deployment. It is very easy to do and runs effectively, even on synthetic data. Resources are available via the OHDSI ATLAS workgroup. While it is a handy complement to the Enclave, do focus on the N3C products: The N3C concept set library and the concept set editor. The Enclave builds its own OMOP vocabulary releases by downloading the vocabulary tables from ATHENA.
Because of these limitations, also do beware that ATLAS “record counts” (the number of patients with data expressing the given concept id) are not linked to N3C and do not represent the counts or distribution of concept ids within the Enclave.
7.3.2 Concept Set Metadata
The work on concept sets does not take place in a vacuum. It is assumed that you are part of a project team, which has the following Subject Matter Expertise (SME): Research/Clinical, Vocabulary, Informatics, and Statistical/Analytic. Each of these SMEs will be called upon in your work.
Intention, Limitations, and Provenance comprise concept set metadata.
7.3.2.1 Intention
Intention communicates in sentences more than the name can. For instance, is the concept set intended to be “broad”, and sensitive, to capture as many cases as possible, leaving downstream analysts to winnow the set of patients down? Or is it “narrow”, and specific? Is it meant to be definitional (these codes “mean” diabetes) or indicative (the codes tell me that you have diabetes, e.g., “retinopathy due to diabetes mellitus”; the codes suggest chronic lung disease, e.g., “infertility due to cystic fibrosis”). “Intention” can also indicate whether the concept set has clinical manifestations or not (e.g., “Sickle Cell” indicates a genetic condition, but unclear if it’s homozygous or heterozygous, the former being clinical and the latter often not). It is true that a later analyst will look through the list of codes, but having an explicit intention helps that analyst screen potential concept sets.
7.3.2.2 Limitations
Limitations communicate edge cases and caveats to the analyst. “Issues” communicates performance with the Enclave data. This performance could include the number of codes contributing the majority of the data (e.g., from Term Usage) or the distribution of values across sites, in the case of lab tests.
7.3.2.3 Provenance
Provenance communicates how the concept set was assembled. It should include any authorities consulted (the literature, the Value Set Authority Center, the domain team members, etc.) and a sense of what modifications, if any, were made from that authoritative beginning. (Of note, “ATLAS” is not an authoritative beginning, unless there is annotation on the ATLAS site of the authoritative source for that specific concept set.)
7.3.3 N3C Concept Set Library
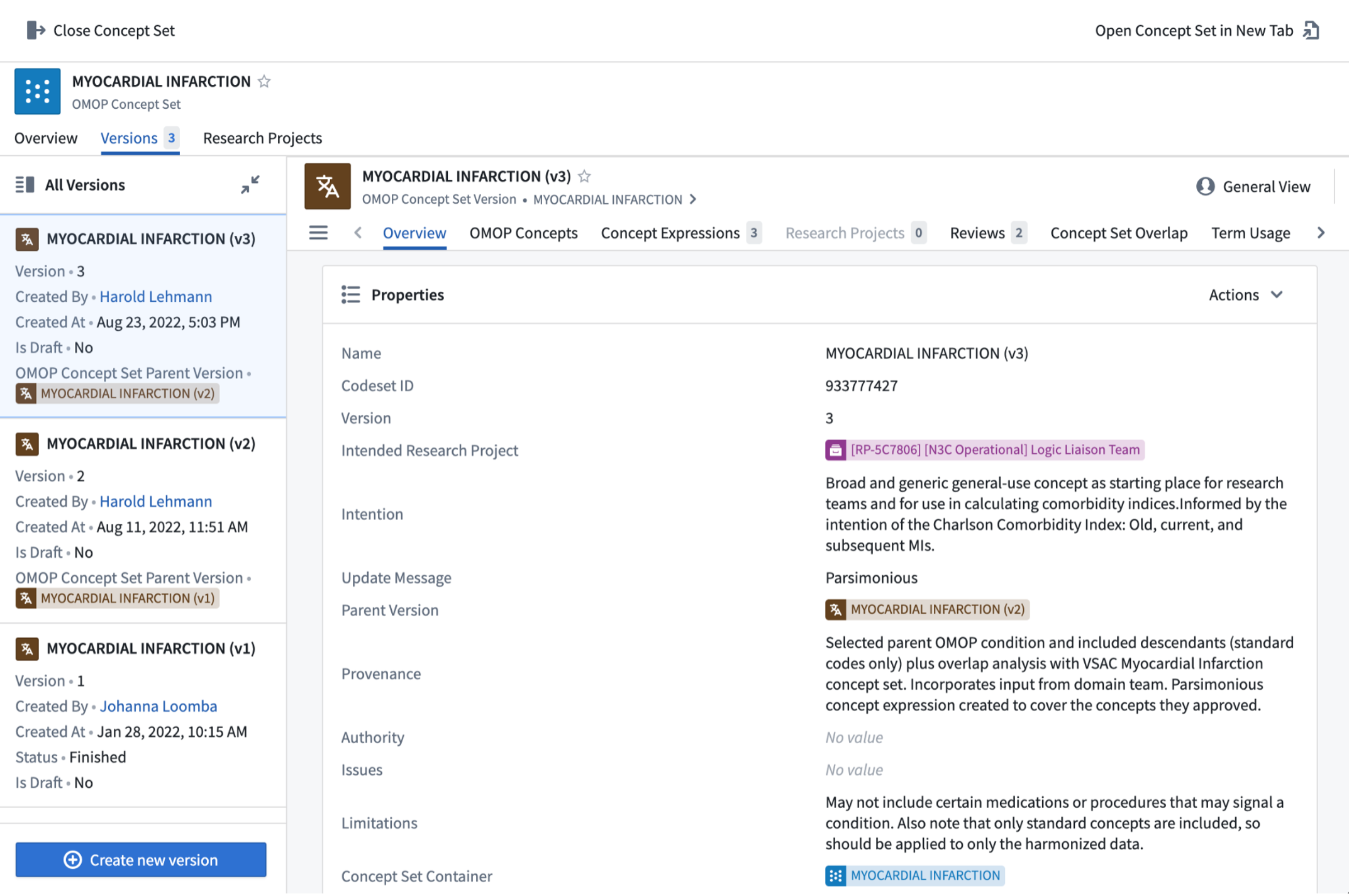
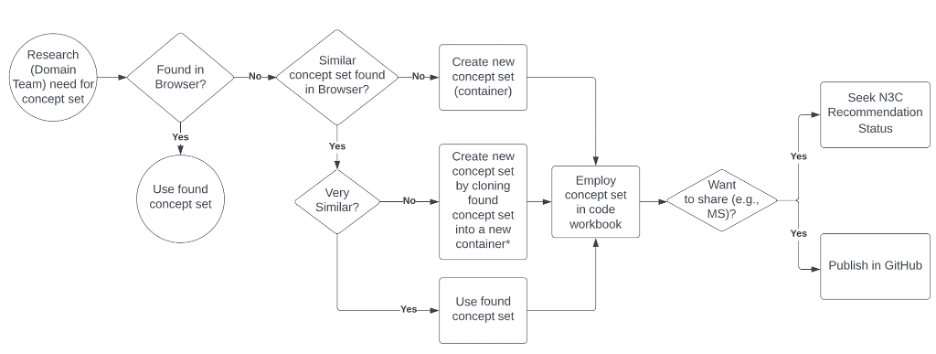
The concept sets already created in the Enclave can be browsed in the concept set browser , illustrated in Figure 7.8. This is the first step for deciding whether a new concept set is needed or if an existing concept set can be chosen for inclusion into an analysis as a building block for an analytic variable or cohort definition. We recommend that you use or build from existing concept sets, especially the N3C Recommended concepts sets, if they serve your research question. A workflow for performing this series of tasks is shown in Figure 7.8.
When created from scratch, a container is created, so multiple versions of the concept sets can be supported. New versions are created as the team gets better facility with the tool, as the team understands better from domain experts subtleties in the intention of the concept set, or as changes are made to the vocabularies, by OHDSI or the source vocabulary stewards. Containers are indicated by blue icons; versions, by brown icons, as illustrated in Figure 7.8.
*New container name should be similar to source container, but different enough to communicate altered intention. MS=Manuscript
The browse screen allows you to filter to concept sets relevant to you. Once you have identified a concept set that you might use, drilling into it provides data about that concept set: versions, metadata, concept expressions, concepts. In reviewing the concept set (generally by drilling to the most recent version), it may be worth exploring similar concept sets using the concept set Overlap tab. At this point, you may be satisfied that an existing concept set serves your needs. If not, you can create a new one – either an entirely new concept set, or a new version of an existing concept set, modifying it to meet your requirements. If you are creating a new version within an existing container, please contact the concept set owner to ensure that your update is in the right spirit. If not, clone the existing version into a new container, naming the container with a name that connotes the difference between the existing and the new concept sets.
If you use a concept set in a published manuscript, make it publicly available. See the next section, Published Concept Sets.
Additional documentation on the concept set browser and editor can be found on the Enclave. A tutorial is available that explores these tools further.
7.3.3.1 Published Concept Sets
“Publication” includes 2 ways of making a concept set available to other researchers. Provisionally Approved and N3C Recommended are automatically submitted to Zenodo, to generate a permanent DOI URL that can be used in publication or in sharing concept set contents with analysts outside the Enclave. Any study manuscript for publication should ensure that any concept set used is either Provisionally Approved or N3C Recommended. The Zenodo URL should appear in the resulting publication.
The N3C Data Liaisons and Logic Liaisons, in partnership with the Domain Teams, have selected common medical features often required for broad use in research that support the analytics pipelines. The original N3C Recommended concept sets were selected based on
the comorbid conditions identified by the US Centers for Disease Control (CDC) definitions for COVID-19 as increasing risk of severe COVID-19
the comorbid conditions identified in the Charlson Comorbidity Index, and
others as requested by N3C leadership for use in the Logic Liaison templates.
The N3C Recommended concept sets have been vetted by clinical experts (typically N3C Domain Team leads) and the Data and Logic Liaison teams according to the below refinement and finalization process. The latest version of the listed concept sets should always be used, as earlier versions represent iterations and have not passed the final review.
The clinical experts and the N3C Data Liaisons and Logic Liaisons perform the following steps in establishing an intentional N3C Recommended concept set:
- Select relevant parent concepts and their descendants from OMOP standard codes, using Charlson Comorbidity description or the relevant section in the referenced CDC page to establish intention.
- Utilize the OHDSI Atlas tool to explore the OMOP hierarchy to look for other potential parent codes and also to remove child codes that are non-specific to the intended scope of the concept set.
- Compare the draft concept set to other related concept sets, using the Concept Set Overlap feature of the Concept Set Editor. This facilitates a review of any codes that do not overlap with those in value sets from reputable stewards such as the National Library of Medicine’s Value Set Authority Center (VSAC) and the Healthcare Cost and Utilization Project (HCUP) and from existing similar concept sets within the Concept Set Browser. Add and remove concepts (and their descendants) as per recommendations from the clinical experts.
- Reduce the intentional concept set expression as parsimonious as possible, retaining all the approved concepts collected in prior steps.
- Iterate as needed.
- Document metadata (properties) in the Concept Set Editor for each created concept set to include: Intention, Limitations, Provenance, and at least one vocabulary and one clinical review.
- Present for final vetting at the Data Liaison meeting for N3C Recommended designation.
These concept sets are in queue to be published as pdf (Properties) and json (concept ids) in Zenodo, to be available to researchers outside the Enclave. Once published, their Zenodo DOI will be posted to the Zenodo Property of the version that was published. The N3C Recommended concept sets are currently published to the N3C GitHub and are available to researchers outside the Enclave.
The N3C Recommended concept sets listed below are used as the default concept sets in the Logic Liaison templates COVID-19 Diagnosed or Lab Confirmed Patients and All Patients which then populate the N3C Phenotype Explorer and N3C Public Health Dashboards.
7.3.3.2 Concept set reviews and validation
Why should anyone trust a concept set? This trust is needed both within the project team and outside the team. The project team needs to trust a concept set in an analysis; someone outside the team needs to assess its trust in deciding whether to (re)use it in their own analysis.
“Trust” in a concept set derives from a number of factors. First, do we trust the people who assembled the concept set and its process? Second, do we believe that the concept set encodes the concepts we want it to encode, and excludes those we do not? Third, does it perform within the Enclave data the way we expect it to?
Re “the people” and “the process”: The concept set meta-data, especially the identity of the author and reviewers, and the Provenance, provide the necessary information.
Another means of deciding on a level of trust is the Reviews. The Enclave supports two types of reviews.
- “Vocabulary review”: Here, a terminologist or informatician familiar with the OMOP vocabulary has reviewed the process documented (in Provenance) or in real-time (as part of work with the Domain Team) and documents satisfaction with what codes are included and excluded.
- “Clinical review”: Here, a clinical member of the Domain Team or project has reviewed the codes to confirm that they match the “Intention” of the concept set.
A third means of deciding on a level of trust is looking to see what research projects have used the concept set. The more authoritative (and published) those projects are, the more authority they have. Of course, in some cases, a concept set “going viral” may simply be a popularity contest and not an indicator of correctness (false positives). At the same time, linking a concept set to a research project is a manual process, which is often not done (false negatives).
Note that this entire scheme of authority and validity depends on adequate documentation of properties. There are many enticing concept sets that do not have properties filled, reviews performed, nor projects linked. Please ensure that your concept sets are not missing these key components.
Note, as well, that, if you publish a paper, using a concept set, that concept set should be published as well. The Properties and Reviews will be published, whether missing or not.
7.4 EHR-Based Data Beyond OMOP
Data tables of use or interest to analysts found in either the LDS or Safe Harbor Release are the following:
7.4.1 Manifest
This table, found in the folder along with the OMOP tables, provides data about each data partner that analysts should find helpful when trying to understand variation in the data: CDM type (ACT, OMOP, PCORNet, TriNetX) and version, whether dates are shifted (and how much), and date of last contribution.
7.4.2 Control Map
For every “case” generated locally by the Phenotype algorithm, the algorithm also identifies 2 “controls”, that are matched by age, sex, race, and ethnicities, including matched by missing values. While it may be tempting to use this map to identify controls for your own study, too many patients who were initial “controls” may have become “cases”, or their true status is unknown, because of home or other non-site testing. Other matching methods (e.g., propensity scores) are more trustworthy.
7.4.3 Microvisit to Macrovisit Map
While admissions to the hospital are recorded in the Visit_Occurrence table, the end date is not always so recorded. Now, in many hospitals, procedures performed during a hospitalization may be recorded in the EHR as an “encounter”. So an admission may be represented in the visit_occurrence table as a string of such “encounters”. We define a macrovisit as a merge of chronological, overlapping inpatient and other longitudinal facility visits, to which we add any other types of visits (outpatient, telehealth, etc) that occur during the merged interval. See the N3C Data Enclave .
7.4.4 Harmonized values
It is no surprise that, across over 70 data partners, and the many included sites, different institutions measure the same analyte in different ways and report the results with different units of measure. N3C has spent a fair amount of effort to harmonize units (and therefore values; think degrees Fahrenheit and degrees Celsius), so analysts have a level playing field for their analyses. In addition, many units of measure are missing, so, as part of that effort, an algorithm has been developed to use the pooled data across the Enclave to help us infer units (and therefore, values). This strategy has led to the “rescue” of over 78% of records where units of measure were missing; see Bradwell et al. (2022). This harmonization was done for about 70 measurements so far. The results are found in the Measurement table, as harmonized_value_as_number, harmonized_unit_concept_id and unit_concept_id_or_inferred_unit_concept_id.
7.4.5 Phenotype and cohort
A phenotype is a grouping of related terms, or observable characteristics that could be applied to a person, disease trait, medical condition or events (Richesson et al., 2013). Examples would be, “has Acute COVID”; “treated with ampicillin”.2
A cohort is a set of persons who satisfy one or more inclusion criteria for a duration of time. Data ingestion is the process of importing data meeting a phenotype definition into a database which can then be used for research. In this section, we will discuss existing tools and methodologies for identifying codes and terms, creating phenotypes, managing vocabulary concept sets, and how to ingest codes into the Enclave database that will enable research using the N3C basic and supplemental datasets.
7.4.6 Derived variables/facts
Facts are designations that a patient falls into a category (at a point in time). Thus, confirmed_covid_patient or TOBACCOSMOKER_indicator is set as “1” (true) if certain conditions are met in the template Logic Liaison Template All Patients Facts . Other Logic Liaison Templates include many derived facts of value to analysis. See Chapter 8.
7.5 Beyond EHR: PPRL-Based Data
7.5.1 Introduction to PPRL
Privacy Preserving Record Linkage (PPRL) is a cryptographically-secure method to link information about individuals from different sources of data, without revealing personal information about those individuals. PPRL enables N3C to collect information such as EHR data from clinical data partners, viral variant information from sequencing centers, mortality information from government and private sources, and government Medicare and Medicaid data, all into a single unified database for researcher use.
7.5.2 PPRL Data Access
Access to datasets that incorporate the PPRL are available through the N3C Data Use Request process. PPRL requests are treated as level 3 / Limited Data Set requests and as such require a signed institutional Data Use Agreement (DUA), a local letter of determination, investigator attestation to the N3C Code of Conduct, IT security training, and human subject training. Requests for use of datasets linked by the PPRL require approval by the National Center for Advancing Translational Sciences (NCATS) federally staffed Data Access Committee (DAC).
7.5.3 Mortality
7.5.3.1 Mortality Data Within N3C
To provide more complete information on mortality, N3C has collected additional mortality data from multiple sources. Because the data sources themselves are sensitive information, supplemental PPRL mortality records are broken down into three source categories:
- Government Mortality: Government data sourced from death certificates and person reporting.
- Public Obituary: Obituary data sourced from funeral homes, newspapers, and other online obituary sources specifically from https://www.obituarydata.com/ (a private obituary aggregator).
- Private Obituary: Obituary data sourced from funeral homes, newspapers, and other online obituary sources sourced from other private sources.
PPRL mortality data are harmonized against the existing Level-3 (LDS) OMOP tables in the N3C Data Enclave. Once access is granted via a Level 3 DUR requesting mortality data, PPRL tables may be found in the Data Catalog under the PPRL Datasets collection. The primary mortality data are available in a table simply called mortality.
7.5.3.2 Mortality Data Completeness and Caveats
First, note that mortality information is available for only those data partners who have opted in to linkage of their records against the mortality data. As a result the data_partner_id column will only represent a subset of the data partner IDs found in other N3C OMOP tables. See the Intro to PPRL documentation for more information.
Secondly, these data sources should not be considered comprehensive in the sense of providing full information on all deaths in the US. As a result, there may well be mortality records in the OMOP death table that are not represented in the supplemental PPRL data. (And certainly there are mortality records in the PPRL data that are not present in the OMOP death table-that’s why N3C has collected this data in the first place!)
Different data sources lag in inclusion of mortality information from the actual date of death (e.g., if someone’s date_of_death is 2022-04-05, that record may not show up in the mortality source data until 2022-04-18). This lag varies by data source type: government sources tend to lag longer than private sources. For detailed information on mortality data latency, data completeness, and other considerations, see the N3C PPRL Mortality Data Guide and the N3C Mortality Data FAQs.
7.5.4 Viral Variant
7.5.4.1 What Are Viral Variants?
Several coronavirus variants have emerged as the virus, SARS-CoV-2, continues to mutate and evolve. For general information about COVID viral variants, see COVID Variants: What You Should Know
7.5.4.2 Viral Variants Data Within N3C
The collection and linkage viral variant PPRL data within N3C is planned in two phases; in the first phase (completed), N3C links patient summary information about sequenced variants. In the second phase (in development), N3C will provide information on the viral variant’s genome sequence via connection to NCBI sequence databases.
For the latest information about viral variant data within N3C, see the N3C PPRL Viral Variants Guide and N3C Viral Variant FAQs.
7.5.4.3 Which sites
The initial viral variant data set includes data from a small number of sites. The number of viral variant data sets will increase as more sites from Clinical and Translational Science Awards (CTSA) and Clinical Trials Research (CTR) participate.
7.5.4.4 Viral Variants Completeness and Caveats
The summary data currently available in N3C includes:
- Sample Date
- WHO label (e.g., alpha, delta, omicron)
- PANGO lineage (e.g., B.1.1.7)3
7.5.4.5 Viral Variant Data in OMOP
Summary PPRL viral variant data is available as an enriched OMOP measurement table available in the PPRL Datasets data. The following columns are used:
measurement_concept_idcontains OMOPconcept_id36033667, “SARS-CoV-2 (COVID-19) variant [Type] in Specimen by Sequencing”, for all viral variant samples.value_as_concept_idwill be encoded with the OMOP concept representing the WHO label (e.g.,concept_id4228611 for “Omicron”).- A new column outside the standard OMOP schema includes the PANGO lineage.
7.5.5 Centers for Medicare and Medicaid Services (CMS)
7.5.5.1 Introduction to CMS
The Centers for Medicare and Medicaid Services (CMS) is a federal agency that is part of the U.S. Department of Health and Human Services (HHS) that administers the nation’s Medicare Program. The agency also works with state governments to administer programs that include Medicaid and the Children’s Health Insurance Program (CHIP).
Medicare is the federal health insurance program created in 1965 to provide health coverage for Americans aged 65 and older. The program was expanded in 1972 to cover people younger than 65 who have permanent disabilities. More than 64 million Americans are currently covered by Medicare.
Medicaid was signed into law in 1965 alongside Medicare. All states, the District of Columbia, and the U.S. territories have Medicaid programs designed to provide health coverage for low-income people. Although the Federal government establishes certain parameters for all states to follow, each state administers its Medicaid program differently, resulting in variations in Medicaid coverage across the country.4
For a full description of CMS, see the HHS website.
7.5.5.2 CMS Claims Data
Medical providers (including pharmacies) submit claims for services or drugs for reimbursement of the enrolled participants. The CMS claims provide an audit trail of billing and reimbursement for healthcare services provided to a patient enrolled in an insurance benefit package no matter where the services were rendered. CMS data contain claims from all sites a patient obtained billable services while enrolled and all medications a patient had filled. CMS claims do not contain clinical depth of EHR data and may not contain non-billable (unbillable) services.
7.5.5.3 CMS Data in N3C
The N3C CMS dataset contains de-identified billing data from the Centers for Medicare and Medicaid Services, formatted into the OMOP common data model for N3C researcher use. The initial CMS data within N3C contains over 240K COVID-19 patients and is updated on a monthly basis. The amount of CMS patients available in the Enclave is dependent on sites participating in the NCATS PPRL linkage initiative, and as additional sites implement the tokenization process the number of patients will increase.
CMS records available cover the period from Jan 1, 2017 to the most recently available (three months ago observation period). Although N3C refreshes its CMS data feed on a weekly basis, CMS data has a significant lag, with most records taking six months or more to become available in the system. All seven types of MS claims are included: Part B (PB), Durable Medical Equipment (DME), Inpatient (IP), Outpatient (OP), Skilled Nursing Facility (SNF), Home Health Agency (HHA), Hospice. The records contain masked providers of service and masked care sites linkable to other characteristics.
The Master Beneficiary Summary File (MBSF):
- Provides enrollment period and reasons for enrollment
- Identifies patients with dual enrollment in Medicare and Medicaid
- Can be used to distinguish non-users from non-eligible
- Identifies periods of continuous enrollment
- Can be used to perform annualization
7.5.5.4 CMS into OMOP CDM Structure
The N3C Data Ingestion and Harmonization (DI&H) team worked with partner Acumen to identify CMS data to be ingested. DI&H then created the mapping from native CMS data elements into the OMOP data model. DI&H created a Code Map Service and a Data Transformation Pipeline, which involved combining sometimes up to 45 rows of data into a single patient record and creating constructs, such as coherent visits across claims. The pipeline also required deidentification of patient and provider identifiers.
7.5.5.5 CMS Data Completeness and Caveats
CMS claims data provides slightly different information compared to EHR data; for example, rather than providing medication prescription information, CMS provides information on medication dispensation from the pharmacy. Enrollment in Medicare and Medicaid are well defined and thoroughly tracked, providing insight into patients healthcare utilization vs eligibility that may be lacking in EHR data.
| EHR Data | CMS Claims Data |
|---|---|
|
|
7.5.5.6 Additional CMS Resources
For a detailed investigation of the usefulness of EHR vs administrative claims data, see Kharrazi et al. (2017).
For detailed information on the N3C CMS data, see N3C PPRL CMS Data Guide .
CMS Research Data Assistance Center (ResDAC) contains data dictionaries, code books, and enormous amounts of information about Medicare claims and enrollment. It is a good place to figure out what values represent. Look for dictionaries for the CMS Standard Analytic Files (SAF) or Limited Data Sets (LDS).
A detailed CMS training webinar is available on YouTube.
7.6 Public/External Datasets
Because our data are not representative of the geographic locations whence they come, it is important for many analyses to attempt to “correct” the results due to this selection bias. Data sets, called external datasets, are available outside the N3C Data Enclave that provide information about such locations. There are datasets about the locations themselves (e.g., zip code distances ), about the demographics in those locations (e.g., zip code census ), or about COVID, in those locations (e.g., COVID-19 vaccine hesitancy in the US by county, state policy by date ). There are also datasets to help in mapping from zip codes (data available in Level 3 Enclave datasets), such as Mapping Zip codes to states and geolocations . All available datasets are available at The Data Discovery Engine (outside the Enclave) and in The Knowledge Store (inside the Enclave; filter on “External Dataset”).
Note that most of these depend on 5-digit zip codes for linking between a patient’s record and the external dataset, and so are useful only in the context of a Level 3 DUR.
Importing such an external dataset entails a process of review that examines the need, the risk to reidentification, and the license. The External Datasets page gives more detail, including how to request a new dataset.
This concludes an exciting chapter in the never ending story of N3C. To a cinema near you. Real soon.
This chapter was first published May 2023. If you have suggested modifications or additions, please see How to Contribute on the book’s initial page.
This translation is automated with software, as opposed to manually performed by a human.↩︎
A phenotype is general and a cohort is specific. Thus, a new-onset diabetes phenotype might say, “Must have some number of outpatient visits without a diabetes diagnostic code, followed by at least one visit with such a code”. A related cohort would be, “Must have 2 outpatient visits without diabetes since Jan 1, 2017 without a diabetes diagnostic code, followed by at least one visit with such a code before Jan 1, 2020”. However, in OHDSI the two terms are used interchangeably. See the Cohorts chapter in OHDSI (2019).↩︎
Note that while WHO label should always be available, PANGO lineage may only be available for a subset of samples↩︎
See https://www.medicareresources.org/glossary/centers-for-medicare-and-medicaid-services/.↩︎